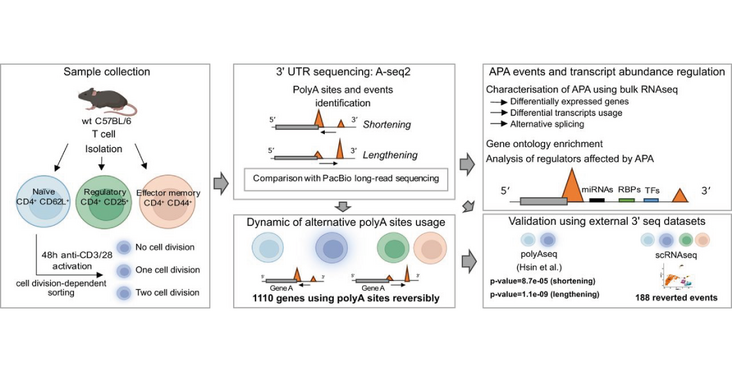
The Jeker and Zavolan labs assessed the 3'-UTR landscape from naive, activated, memory and regulatory CD4+ cells. After activation, the T cells undergo a rapid expansion, and during this period alternative polyadenylation events occured, which reverted back after the expansion phase. These changes did not seem to correlate with transcript abundance. However, the alternative 3'-UTRs contained binding sites for miRNAs and RNA binding proteins, known to be important for T-cells which could imply alternative polyadenylation in the control of cell fate and homeostasis. Their findings were published in the article "T helper cells exhibit a dynamic and reversible 3'-UTR landscape" in the RNA journal.
Abstract
3' untranslated regions (3' UTRs) are critical elements of messenger RNAs, as they contain binding sites for RNA-binding proteins (RBPs) and microRNAs that affect various aspects of the RNA life cycle including transcript stability and cellular localization. In response to T cell receptor activation, T cells undergo massive expansion during the effector phase of the immune response and dynamically modify their 3' UTRs. Whether this serves to directly regulate the abundance of specific mRNAs or is a secondary effect of proliferation remains unclear. To study 3'-UTR dynamics in T helper cells, we investigated division-dependent alternative polyadenylation (APA). In addition, we generated 3' end UTR sequencing data from naive, activated, memory, and regulatory CD4+ T cells. 3'-UTR length changes were estimated using a nonnegative matrix factorization approach and were compared with those inferred from long-read PacBio sequencing. We found that APA events were transient and reverted after effector phase expansion. Using an orthogonal bulk RNA-seq data set, we did not find evidence of APA association with differential gene expression or transcript usage, indicating that APA has only a marginal effect on transcript abundance. 3'-UTR sequence analysis revealed conserved binding sites for T cell-relevant microRNAs and RBPs in the alternative 3' UTRs. These results indicate that poly(A) site usage could play an important role in the control of cell fate decisions and homeostasis.
Read the Article in RNA (Open Access)
Abstract, figure and title from Seyres, Gorka, Schmidt et al. (2024) RNA published under a CC BY 4.0 license.