Ciliates represent a unique system to study nuclear biology, as they have a micro- and a macronucleus. Formation of the macronucleus requires the excision of thousands of transposon related sequences, which is guided by small RNAs. The Nowacki lab could now show that in ciliates the excision of these DNA sequences also requires the activity of the ciliate chromatin remodelling factor ISWI1, which is a homolog of ISWI in other organims inlcuding metazoans. Also, they could identify co-factors of ISWI1 required for this process. Their findings were published in the EMBO Journal in the article "Chromatin remodeling is required for sRNA-guided DNA elimination in Paramecium".
Synopsis
Small non-coding RNAs regulate genome reorganization in the ciliate Paramecium in a tightly controlled fashion. This study reports cross-talk between a chromatin remodeler, ISWI1, and the sRNAs-associated argonaute proteins Ptiwi01 and Ptiwi09 for the precise excision of transposable elements by the transposase PGM and genome reorganization during sexual development.
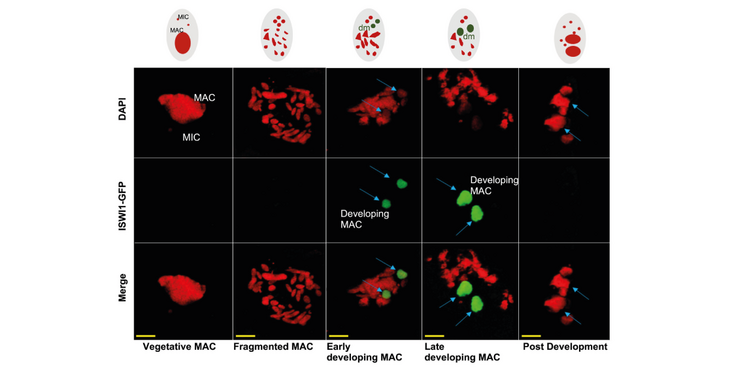
- Paramecium ISWI homolog ISWI1 is differentially upregulated during macronuclear development and affects DNA elimination.
- ISWI1 knockdown leads to alternative excision of IESs, including excision at the “forbidden peak” ~35?bp from the IESs, a region which is excluded from excision in wildtype.
- Ptiwi01 and Ptiwi09 interact with ISWI1 during sexual development.
- Knockdown of ISWI1 or the transposase PGM alters the distribution of nucleosome densities.
Abstract
Small RNAs mediate the silencing of transposable elements and other genomic loci, increasing nucleosome density and preventing undesirable gene expression. The unicellular ciliate Paramecium is a model to study dynamic genome organization in eukaryotic cells, given its unique feature of nuclear dimorphism. Here, the formation of the somatic macronucleus during sexual reproduction requires eliminating thousands of transposon remnants (IESs) and?transposable elements scattered throughout the germline micronuclear genome. The elimination process is guided by Piwi-associated small RNAs and leads to precise cleavage at IES boundaries. Here we show that IES recognition and precise excision are facilitated by recruiting ISWI1, a Paramecium homolog of the chromatin remodeler ISWI. ISWI1 knockdown substantially inhibits DNA elimination, quantitatively similar to development-specific sRNA gene knockdowns but with much greater aberrant IES excision at alternative boundaries. We also identify key development-specific sRNA biogenesis and transport proteins, Ptiwi01 and Ptiwi09, as ISWI1 cofactors in our co-immunoprecipitation studies. Nucleosome profiling indicates that increased nucleosome density correlates with the requirement for ISWI1 and other proteins necessary for IES excision. We propose that chromatin remodeling together with small RNAs is essential for efficient and precise DNA elimination in Paramecium.
Read the Publication in the EMBO Journal (Open Access)
Abstract, figure, synopsys and title from Singh et al (2022) EMBO Journal published under a CC BY 4.0 license.