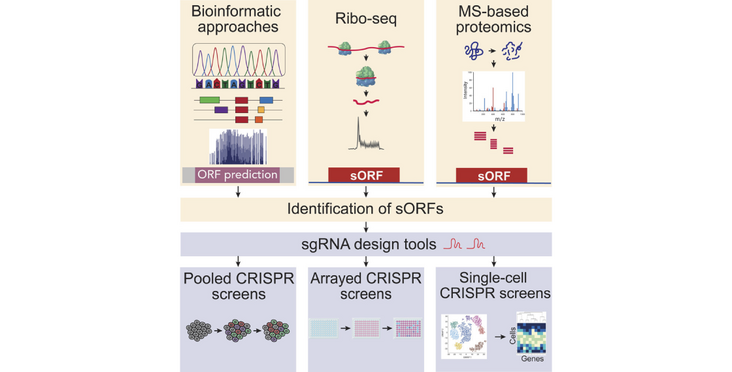
The human genome is estimated to encode in small open reading frames more than 7000 microproteins. Their size makes it necessary to use dedicated approaches to study their functions which are still largely unknown. The Sendoel lab wrote a review on methods & tools to study microproteins entitled "No country for old methods: New tools for studying microproteins" published in iScience.
Abstract
Microproteins encoded by small open reading frames (sORFs) have emerged as a fascinating frontier in genomics. Traditionally overlooked due to their small size, recent technological advancements such as ribosome profiling, mass spectrometry-based strategies and advanced computational approaches have led to the annotation of more than 7000 sORFs in the human genome. Despite the vast progress, only a tiny portion of these microproteins have been characterized and an important challenge in the field lies in identifying functionally relevant microproteins and understanding their role in different cellular contexts. In this review, we explore the recent advancements in sORF research, focusing on the new methodologies and computational approaches that have facilitated their identification and functional characterization. Leveraging these new tools hold great promise for dissecting the diverse cellular roles of microproteins and will ultimately pave the way for understanding their role in the pathogenesis of diseases and identifying new therapeutic targets.
Read the Publication in iScience (Open Access)
Website Sendoel Lab
Abstract, figure and title from Valdivia-Francia and Sendoel (2024) iScience published under a CC BY-NC-ND 4.0 license.