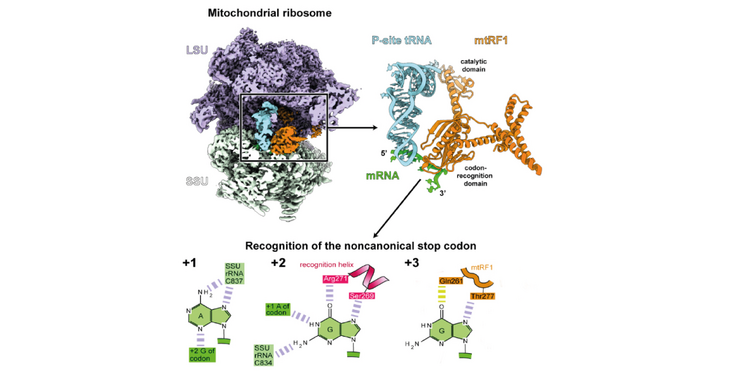
A recent study published in “Science” by the Ban lab (IMBB) together with the Fillipovska (Harry Perkins Institute of Medical Research, Perth, Australia) and Gatfield (University of Lausanne) groups sheds light on the mechanism by which the mitochondrial release factor 1 (mtRF1) recognizes noncanonical stop codons during translation termination.
Vertebrate mitochondrial genomes frequently deviate from the common usage of the genetic code. They use four instead of three stop codons for translation termination: Two of them are canonical stop codons, whereas the third canonical stop codon codes for an amino acid. In addition, two codons that usually specify for amino acids act as noncanonical stop codons.
The protein required for the release of the newly synthesized protein during translation termination at these noncanonical stop codons was previously unknown. Researchers from the Ban, Filipovska, and Gatfield groups used gene-editing and ribosomal profiling in combination with cryo-electron microscopy to identify mtRF1 as the protein responsible for the release of polypeptides at noncanonical stop codons.
The researchers discovered that mtRF1 recognizes noncanonical stop codons by binding to the decoding center of the ribosome, which distorts the mRNA in an unusual conformation. As a consequence, the recognition of the stop codon is not exclusively mediated by specific contacts with the release factor, as it is the case for all standard codons, but through a network of unusual interactions that also include the ribosomal RNA.
This discovery provides insights into a molecular mechanism of a translation termination process that originates from a recoding event in the mitochondrial genome.
Read the Publication in Science