Inhibiting ribosome biogenesis is a promising strategy for treating cancers. Despite the many factors involved that could represent potential drug targets only few inhibitors have been identified to date. To help this aim, the Kutay and Horvath labs have developed an imaging-based screening pipeline with four readouts and counter assays to help exclude hits caused by other targeting mechanisms. Their pilot screen with over 1000 compounds obtained very good quality screening scores. Their findings have been published in the article “Establishment of an imaging-based screening pipeline for the identification of human ribosome biogenesis inhibitors”.
Abstract
Background: Ribosomes are huge ribonucleoprotein particles that mediate protein synthesis in all organisms. The synthesis of ribosomes is a complex process that involves hundreds of supporting factors in mammalian cells, including proto-oncogenes and tumor suppressors. Dysregulation of ribosome biogenesis can contribute to tumorigenesis, and the increased production of ribosomes in cancer cells is known to promote proliferative cell growth. Therefore, ribosome biogenesis represents an attractive vulnerability of cancer cells that ought to be exploited for the development of anti-cancer drugs. Despite the large number of trans-acting factors promoting ribosome assembly including potentially druggable enzymes, only few chemical inhibitors that act on ribosome biogenesis, especially downstream of pre-rRNA transcription, have been identified to date.
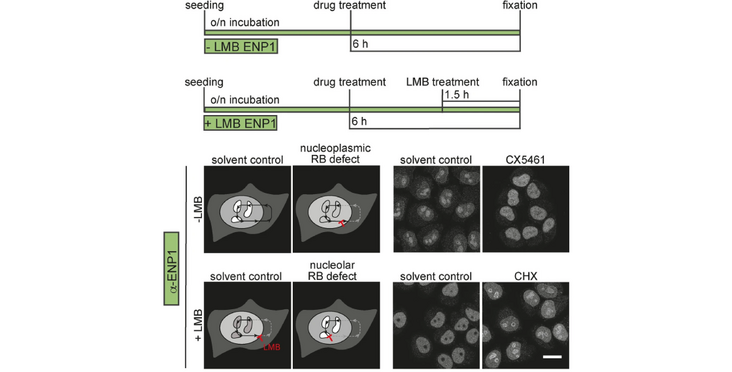
Results: To enable large-scale screens for chemical compounds that interfere with ribosome biogenesis, we have established a pipeline to perform single-cell, imaging-based screening campaigns using four different readouts, including fluorescent ribosomal protein reporters (RPS2-YFP, RPL29-GFP) and immunofluorescence analyses of the ribosome biogenesis factor ENP1(BYSL), in HeLa cells, a human cancer line. We have assessed the robustness of our high-content screening approach by performing a pilot screen using a library comprising more than 1000 FDA-approved drugs with known targets in other pathways. This pilot screen obtained excellent quality scores and identified ten compounds as hits. These hit compounds likely affect ribosome synthesis indirectly, the majority by inducing DNA damage or by inhibiting the proteasome. We therefore used the identified compounds to establish appropriate counter assays for DNA damage and proteasome inhibition, to exclude common indirect effects in the downstream analysis of such screening campaigns.
Conclusions: The established screening pipelines provide a robust, efficient, and sensitive experimental framework to identify chemical compounds that impair ribosome synthesis. The combination of readouts allows to distinguish effects on pre-rRNA synthesis from downstream effects on ribosome assembly. Established counter assays on DNA damage and protein degradation enable to exclude effects on these pathways, which commonly interfere with ribosome synthesis indirectly. The developed assays are easily scalable to screen libraries of higher complexity in the future.
Read the Publication in BMC Biol (Open Access)
Abstract and figure from Gafko (2025) BMC Biol published under a CC BY 4.0 license.