The Pieters and Zavolan Labs developed a novel approach based on single-cell RNA Sequencing to detect biological variation between populations of cells. They used their approach to study the gene expression landscape of unstimulated macrophages derived from two different mouse strains, which have different infection susceptibility. The unstimulated macrophage population from the more pathogen resistant strain, showed a more similar expression profile to stimulated macrophages thant the ones from the more susceptible strain. Their findings were published in the article "A novel approach to single-cell analysis reveals intrinsic differences in immune marker expression in unstimulated BALB/c and C57BL/6 macrophages" in FEBS Letters.
Abstract
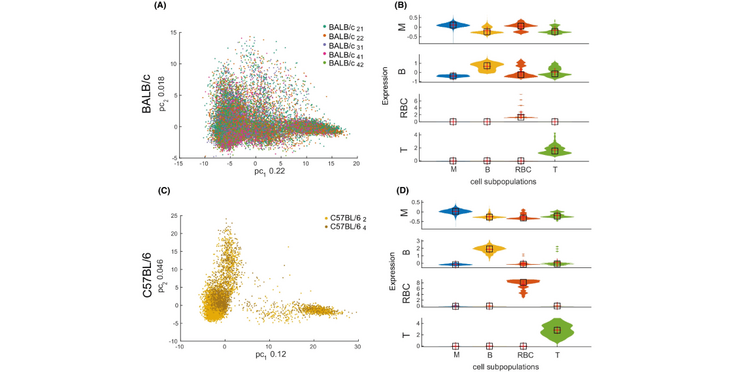
The origin of functional heterogeneity among macrophages, key innate immune system components, is still debated. While mouse strains differ in their immune responses, the range of gene expression variation among their pre-stimulation macrophages is unknown. With a novel approach to scRNA-seq analysis, we reveal the gene expression variation in unstimulated macrophage populations from BALB/c and C57BL/6 mice. We show that intrinsic strain-to-strain differences are detectable before stimulation and we place the unstimulated single cells within the gene expression landscape of stimulated macrophages. C57BL/6 mice show stronger evidence of macrophage polarization than BALB/c mice, which may contribute to their relative resistance to pathogens. Our computational methods can be generally adopted to uncover biological variation between cell populations.
Read the Publication in FEBS Letters (Open Access)
Abstract, figure and title from Breda et al. (2022) FEBS Letters published under a CC BY-NC 4.0 license.