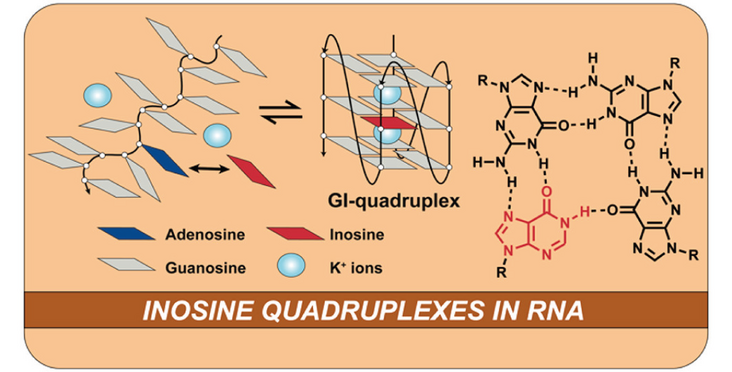
The lab of Jonathan Hall in collaboration with the Allain, Bergmann and Zenobi labs investigated the influcence of Adenosine to Inosine substiutions on RNA structure, which are catalyzed in cells by ADAR enzymes. They found that introduction of inosines leads to the formation of G-quadruplexes termed GI quadruplexes. Their findings have been published in JACS in an article entitled "Inosine Substitutions in RNA Activate Latent G-Quadruplexes".
Abstract
It is well-accepted that gene expression is heavily influenced by RNA structure. For instance, stem-loops and G-quadruplexes (rG4s) are dynamic motifs in mRNAs that influence gene expression. Adenosine-to-inosine (A-to-I) editing is a common chemical modification of RNA which introduces a nucleobase that is iso-structural with guanine, thereby changing RNA base-pairing properties. Here, we provide biophysical, chemical, and biological evidence that A-to-I exchange can activate latent rG4s by filling incomplete G-quartets with inosine. We demonstrate the formation of inosine-containing rG4s (GI-quadruplexes) in vitro and verify their activity in cells. GI-quadruplexes adopt parallel topologies, stabilized by potassium ions. They exhibit moderately reduced thermal stability compared to conventional G-quadruplexes. To study inosine-induced structural changes in a naturally occurring RNA, we use a synthetic approach that enables site-specific inosine incorporation in long RNAs. In summary, RNA GI-quadruplexes are a previously unrecognized structural motif that may contribute to the regulation of gene expression in vivo.
Read the Article in JACS (Open Access)
Abstract, figure and title from Hagen et al. 2021 JACS published under a CC BY-NC-ND 4.0 license.