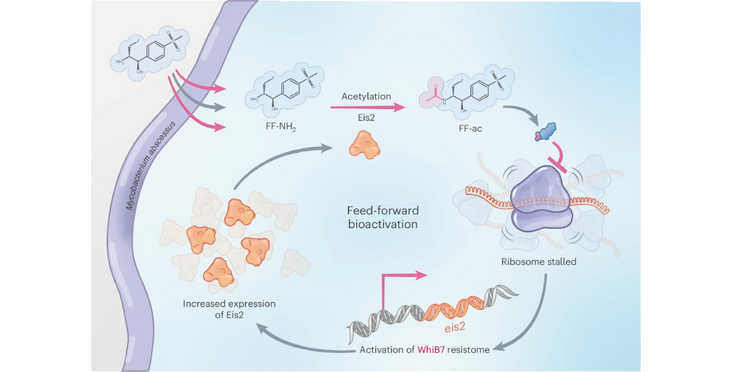
Non-tuberculous mycobacteria are an emerging health threat and are difficult to treat due to their high resistance to antibiotics. Mycobacterium abcessus causes particularly difficult to treat infections with an estimated mortality of 50% over 15 years, which has caused M. abcessus to be called an “antibiotic nightmare”.
The Lee and Sander labs (associate member of the NCCR RNA & Disease) have now discovered a potential treatment strategy based on a pro-drug which is converted by a feed-forward loop to its active form by an enzyme belonging to the central resistance pathway of M. abcessus. The drug then inhibits the mycobacterial ribosome. Bacteria have been observed to develop resistance to this compound by either mutating the conversion enzyme or the controlling transcription factor for resistance. Mutations in these proteins render the bacteria then susceptible to antibiotics they have been previously resistant to.
The new compound showed efficacy in a murine model of M. abcessus infection, a narrow spectrum activity against M. abscessus-chelonae complex species and no toxicity e.g. associated with inhibiting mammalian mitoribosomes. The authors propose an alternating treatment scheme with the new compound and established antibiotics.
Read the Publication in Nature Microbiology (Open Access)
Figure from Phelps, Kurt et al. (2025) Nature Microbiology published under a CC BY 4.0 license.