The Polacek lab in collaboration with the Hapfelmeier group found that the small stationary phase-specific RNA FimR2 in Escherichia coli and its homoglogue in Salmonella enterica are regulators to environmental conditions. They are involved in controlling various processes such as synthesis of fimbria and flagella and protein secretion regulating thereby bacterial mobility, biofilm formation and virulence. Their findings have been published in the article "The stationary phase-specific sRNA FimR2 is a multifunctional regulator of bacterial motility, biofilm formation and virulence " Nucleic Acids Research.
Abstract
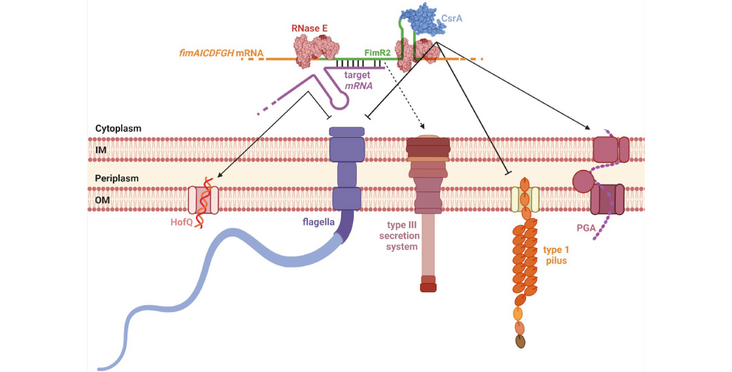
Bacterial pathogens employ a plethora of virulence factors for host invasion, and their use is tightly regulated to maximize infection efficiency and manage resources in a nutrient-limited environment. Here we show that during Escherichia coli stationary phase the 3' UTR-derived small non-coding RNA FimR2 regulates fimbrial and flagellar biosynthesis at the post-transcriptional level, leading to biofilm formation as the dominant mode of survival under conditions of nutrient depletion. FimR2 interacts with the translational regulator CsrA, antagonizing its functions and firmly tightening control over motility and biofilm formation. Generated through RNase E cleavage, FimR2 regulates stationary phase biology by fine-tuning target mRNA levels independently of the chaperones Hfq and ProQ. The Salmonella enterica orthologue of FimR2 induces effector protein secretion by the type III secretion system and stimulates infection, thus linking the sRNA to virulence. This work reveals the importance of bacterial sRNAs in modulating various aspects of bacterial physiology including stationary phase and virulence.
Read the Publication in Nucleic Acids Research (Open Access)
Abstract, figure and title from Raad et al. (2022) Nucleic Acids Res published under a CC BY-NC 4.0 license.