NMD-sensitive transcripts catalogued at unprecedented detail in human cells
Cells harbour a variety of quality control mechanisms to prevent the production and accumulation of harmful proteins. The quality control mechanism nonsense-mediated mRNA decay (NMD) is a translation-dependent RNA degradation pathway that targets mRNAs with premature stop codons as well as many mRNAs that encode full-length proteins. NMD is essential for correct embryonic development in all mammals and plays an important role in tumors and genetic diseases such as cystic fibrosis. In a new study published in the journal Genome Biology, the labs of Oliver Mühlemann (University of Bern) and Mihaela Zavolan (Biozentrum in Basel) joined forces to gain new insights into this quality control mechanism by mapping endogenous NMD targets on a global scale at unprecedented detail using Nanopore sequencing (Karousis et al., Genome Biology, 2021).
NMD was initially discovered to degrade aberrant mRNAs that carry premature termination codons (PTC) arising from mutations or errors in RNA processing, such as transcription or splicing. Later studies that characterized NMD substrates on a transcripome-wide scale revealed that NMD not only removes PTC-containing transcripts, but also controls the overall expression level of many mRNAs encoding full-length proteins. These studies sparked the efforts to understand what features of transcripts beyond the presence of an early stop codon render transcripts NMD-sensitive.
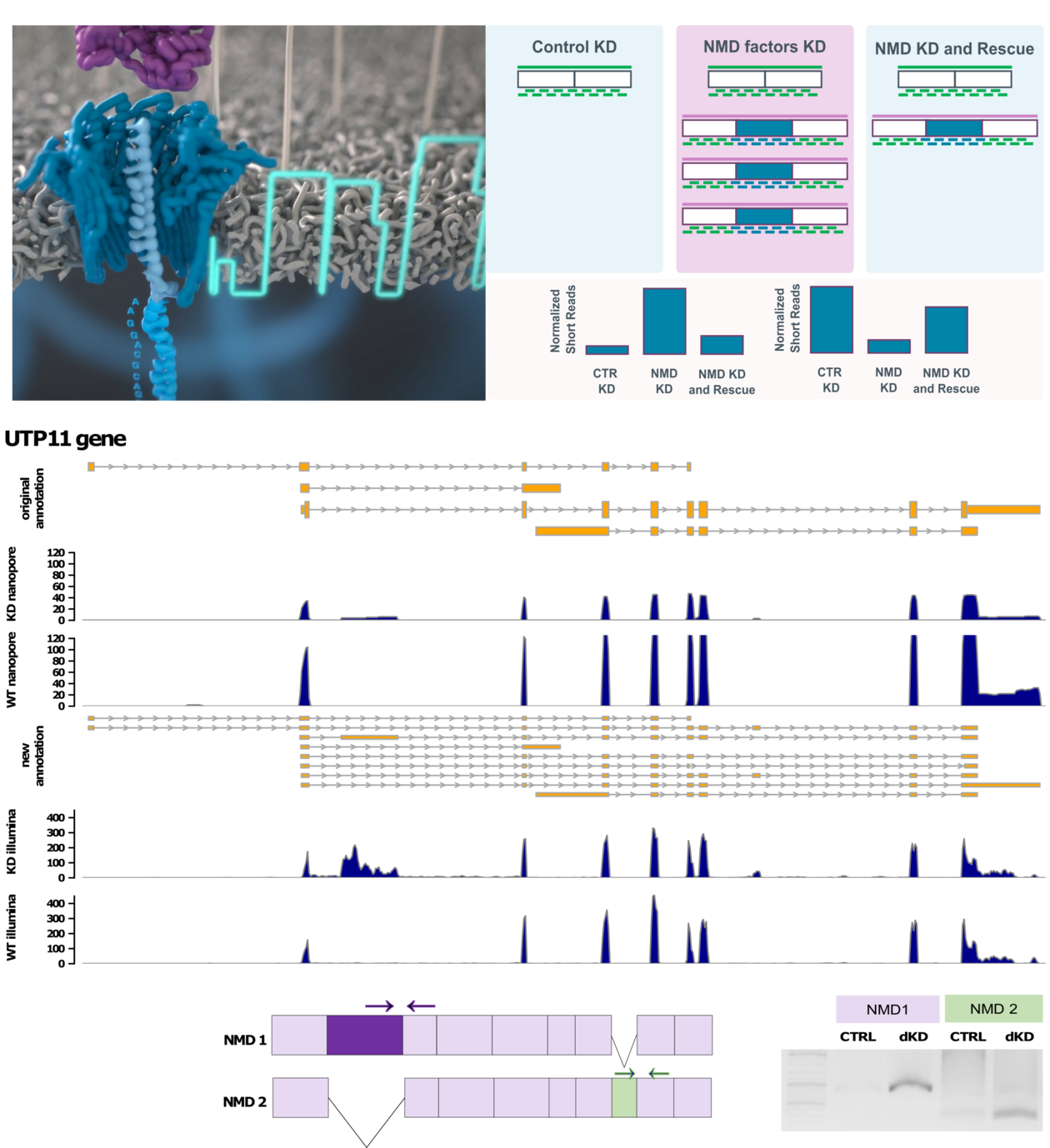
In healthy cells, where NMD is active, it is difficult to detect defective mRNAs because they are rapidly degraded after production. To detect NMD targeted transcripts, Karousis and colleagues depleted the key NMD factors UPF1, SMG6 and SMG7 either individually or in combination in HeLa cells. Under these conditions, NMD-sensitive transcripts accumulate and can be detected by high-throughput sequencing.
Previous methods for detecting NMD-targets in cells have relied on short-read sequencing techniques (Illumina) generating short sequences that are later computationally reassembled into whole transcripts. However, in this gigantic puzzle, many pieces cannot be unambiguously assigned to a specific transcript. Evan Karousis, first author of the study, explains that they applied Nanopore sequencing - a rather new method that decodes full-length mRNAs in one piece. "This allows to overcome the limitation of short-read sequencing technologies as each detected mRNA can uniquely be assigned to an mRNA variant", Karousis said. “In addition, Nanopore sequencing allows for the discovery of new RNA isoforms, among which many are NMD targets”. The combination of long-read Nanopore sequencing with short-read Illumina sequencing enabled the researchers to create a comprehensive and high-fidelity catalog of NMD targeted RNAs in human cells.
Using this catalog of NMD-sensitive transcripts, the researchers tried to identify features that would allow ab initio prediction of NMD targets. They could confirm a central role of exon junctions in the 3' UTR for rendering a transcript sensitive to NMD. Surprisingly - and in contrast to previous findings -, the length of the 3' UTR did not correlate with NMD sensitivity for mRNAs with the stop codon in the last exon. And finally, the authors showed that NMD targets both canonically and non-canonically spliced mRNAs – supporting the notion that NMD serves not only as a quality control mechanism to remove faulty spliced transcripts but also as a general mechanism of regulating gene expression.
The importance of these findings is evident: The accurate classification of NMD sensitive transcripts and the identification of NMD-inducing features has the potential to provide insights into the degradation mechanism. "If certain mRNAs accumulate that are degraded in healthy cells by NMD quality control, this can contribute to the development of tumors, as is the case in gastric cancer, for example", Oliver Mühlemann explains. So if we understand how the quality control can distinguish defective from correct mRNAs, this knowledge will contribute to the development of new therapeutic approaches for diseases in which quality control is impaired.
Karousis et al. (2021) Genome Biology, 22(1), 223 (Open Access)
Top left image with permission from Oxford Nanopore. Two other figures from Karousis et al. (2021) Genome Biology published under a CC BY 4.0 license.