Sending a second messenger
Since its discovery a few years ago, the CRISPR-Cas system - or rather a simplified version of it (CRISPR-Cas9) - has triggered a biotech revolution due to its use an effective tool to modify the genomes of cells or entire organisms. By delivering the Cas9 nuclease complexed with a synthetic guide RNA into a cell, the genome can be cut at a desired location to remove existing genes and to add new ones. Originally though, CRISPR systems and their associated Cas proteins are found in bacteria, in which they function as an immune system to provide protection against genetic invaders such as viruses and plasmids. In these systems, the invaders are recognized by effector molecular complexes that use short RNA molecules as molecular guides to bind the invader's DNA or RNA and target it for destruction. The exact mechanisms of this prokaryotic immune response are still being investigated but have drawn striking parallels between prokaryotic and vertebrate innate immune systems, as reported recently by an international research team headed by Martin Jinek of the University of Zurich. So CRISPR-Cas systems not only serve as a revolutionary genome editing tools but also prove to be a fascinating field for basic microbiology research. Jinek et al. were able to show a new CRISPR-Cas defense mechanism involving a "second messenger" that is synthesised when the bacterial immune system detects an invading virus. The mechanism was discovered in a close collaboration with the research group of Jonathan Hall from the ETH Zürich, whose expertise in the chemical synthesis and analysis of modified ribonucleic acids has made a critical contribution to the project. The team effort, facilitated by the NCCR RNA & Disease, yielded results that point to an unprec-edented mechanism for regulation of CRISPR immunity.
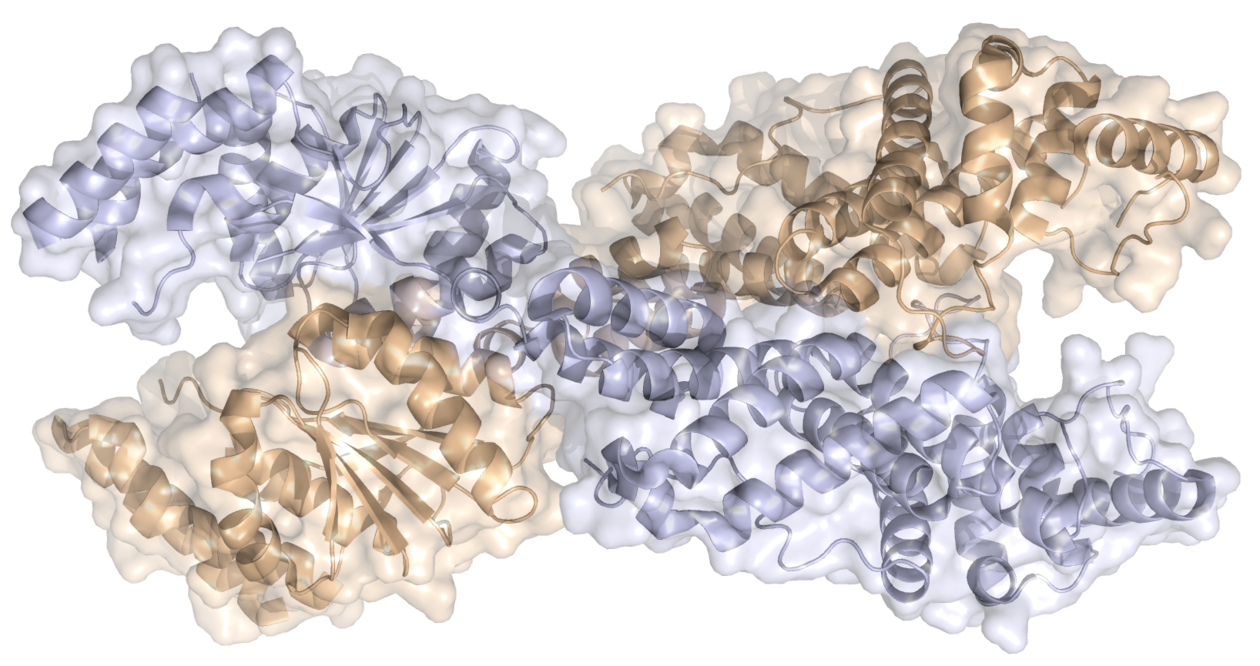
At the heart of the mechanism is a CRISPR-associated protein known as Csm6. It had previously been shown before that the CRISPR-associated protein Csm6 contributes to invader immunity by functioning as a ribonuclease that degrades invader-derived RNAs, but the mechanism linking invader sensing to Csm6 activity was not understood. Jinek's group now shows that Csm6 proteins are activated by a cyclic RNA molecule composed of four or six adenine bases linked in a circular manner (cyclic oligoadenylates). These "second messenger" molecules are synthesised from ATP by the RNA-guided effector complex when it detects an invader RNA. The messengers in turn allosterically activate the Csm6 RNase by binding to its CRISPR-associated Rossmann fold (CARF) domain. As the synthesis of cyclic oligoadenylate is triggered by invader RNA recognition, this provides a failsafe interference mechanism in case the intrinsic DNase and RNase activities of the CRISPR system are insufficient to counteract the invader, such as when the target gene is expressed late during viral infection.
The newly found mechanism for regulation of CRISPR interference is strikingly similar to a well-known mechanism found in mammalian innate immunity in which viral RNAs trigger the production of oligoadenylate second messengers that in turn activate a cellular ribonuclease to promote viral RNAs degradation. "So bacteria, in their own way, fight viral infections in a way that is surprisingly similar to what human cells do", Jinek says. Moreover, the study notes that other CRISPR-associated proteins that are predicted to respond to the second messengers appear to be transcription factors rather than nucleases. This raises the intriguing possibility that CRISPR-Cas systems might also use cyclic oligoadenylate signaling to activate other host genes to help fight genetic invaders.
Niewoehner O. et al. (2017) Nature 548, 543-548
By Roland Fischer