Keeping the shape to cut off-targets
https://doi.org/10.1016/j.cell.2022.09.026 https://creativecommons.org/licenses/by-nc-nd/4.0/
In 2023, the British health authorities and then the FDA approved the first gene editing therapy based on CRISPR-Cas9, developed to treat Sickle Cell Disease and Beta-Thalassemia. These approvals come less than twelve years after it was published that these adaptive immune systems of bacteria could be used for genome editing. A bench-to-bedside development time extremely fast compared to other new biotechnological therapies such as monoclonal antibodies, antisense oligonucleotides and siRNAs. "I feel like I am living a dream in the sense that I have contributed to a scientific discovery that has potential to cure diseases, and then this potential is fulfilled within a decade," states Martin Jinek, who, as a postdoc in the lab of Jennifer Doudna, co-discovered the use of CRISPR-Cas9 as a genome editing tool.
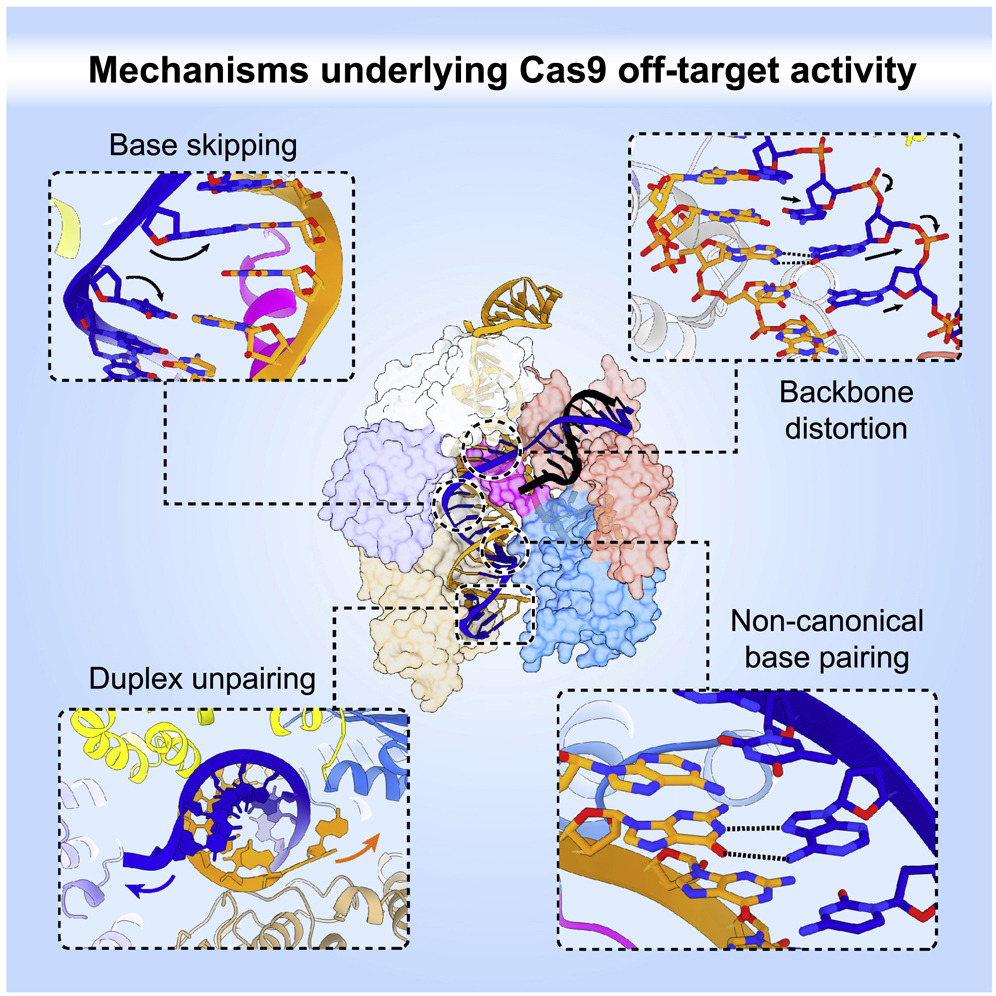
However, fundamental questions such as insights into the mechanism of off-target activity remain, which is an essential topic regarding safety for clinical applications. The Jinek lab (University of Zurich), in collaboration with scientists from the gene editing company Caribou Biosciences and academic researchers from other groups, set out to find the structural basis for the off-target DNA cutting of Cas9, which has been shown to tolerate mismatches between the guide RNA and the target DNA strand. To shed light structurally on the off-target activity of CRISPR-Cas9, the researchers solved and analyzed 19 crystal structures of CRISPR-Cas9 in complex with guide RNA and DNA, of which 16 were off-target complexes selected from a list of in vivo determined off-targets to represent all possible base mismatches and located at different position of the guide target duplex.
The structural data were complemented by molecular dynamics simulations, as well as the determination of the kinetics and thermodynamics of substrate binding and cleavage efficiency. To determine the kon, koff values and binding constant, they applied the switchSENSE technology available through the NCCR's NMR technology platform. This wealth of data allowed the researchers to draw general conclusions on how Cas9 accommodates different types of mismatches.
Cas9 can accept not matching sequences between the guide RNA and the target DNA as long as the overall shape of the RNA-DNA Duplex is not too perturbed. For mismatches, the duplex shape is maintained by the formation of non-canonical base pairs. For deletions, base skipping or multiple mismatches allow for the maintenance of the duplex shape. Interestingly, bulge formation by the guide RNA was not observed. These adaptations for off-target sequences maintained nearly all contacts between Cas9 and the bound duplex. "The system has evolved to be specific enough so that the bacterium does not kill itself by accident, but at the same time, it has a little bit of promiscuity so that it can deal with mutating viruses," states Martin Jinek, last author of the publication in Cell.
Despite maintaining a duplex shape, the positioning of the Cas9 machinery is not optimal anymore for off-targets, which leads, depending on the case, to a slight to very strong reduction in cleavage efficiency and speed. Depending on the off-target, this reduction is due to perturbations to either the formation of R-loops or the conformational activation checkpoint, as well as steric hindrance of the Cas9 HNH domain. "This system is, from a structural viewpoint, incredibly plastic. It is a big combinatorial game, and we have only scratched its surface," says Martin Pacesa, the paper's first author of the paper.
Overall, the researchers' findings, besides shedding further light on the fascinating molecular machine that Cas9 represents, should help create high-fidelity Cas9 variants, aid in guide RNA design and off-target predictions contributing to increasing the safety and specificity of Cas9-based therapies. "Cas9 is an amazing molecular machine - it binds DNA, it binds RNA, it can unwind DNA, it can cleave DNA and undergo several conformational changes and all that without consuming any ATP or any kind of external source of energy. So what one protein can do is quite impressive," concludes Martin Pacesa.
Publication:
Pacesa et al. (2022) Cell 185(22):4067-4081 (Open Access)
Text: Dominik Theler