Surprising insights into UV Cross-Linking of Proteins and RNA
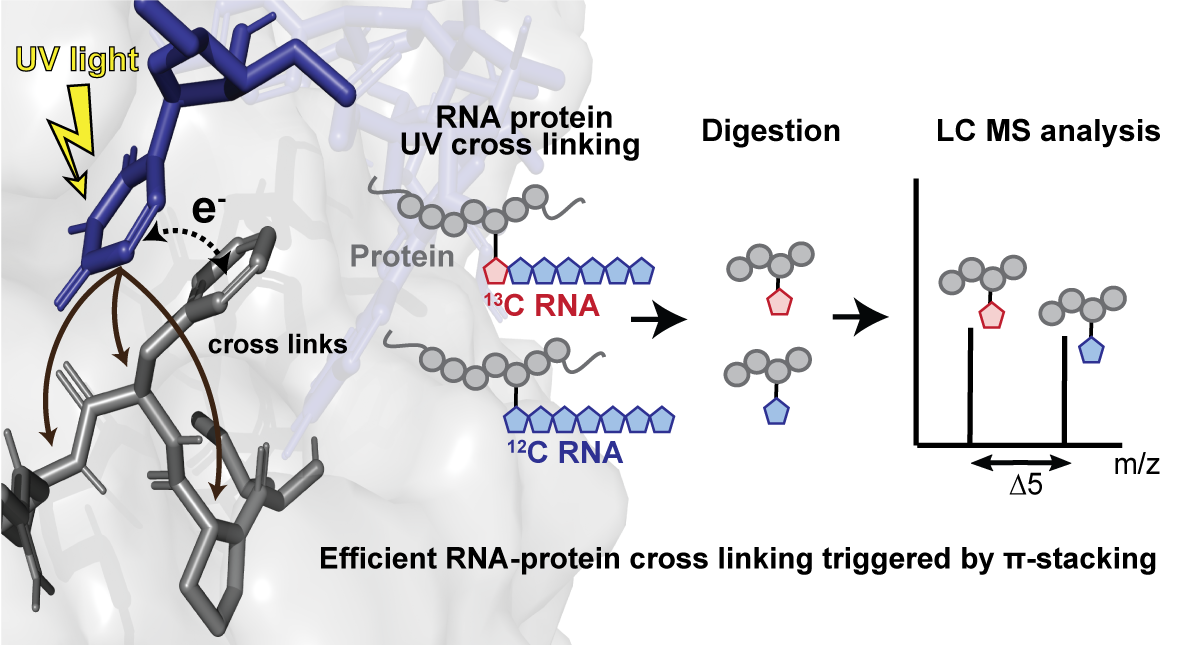
Researching RNA protein interactions at different scales and resolutions often involves UV cross-linking. However, UV-induced cross-linking between RNA and proteins is not a well-understood process despite its widespread use. The lab of Jonathan Hall joined forces those of Ruedi Aebersold, Fred Allain and Alexander Leitner at ETH Zurich to gain insights into this process.
Gaining insights was not as straightforward as it seems. Cross-linking of amino acids and nucleotides requires high local concentrations and the special environment of the protein domain. Hence, it is difficult to mimic the cross-linking reactions under conventional reaction conditions in aqueous solutions. Furthermore, cross-linking often leads to complex mixtures of reaction products that are difficult to characterize.
As a model system for cross-linking studies, the researchers turned to the RNA Recognition Motif (RRM) domain of the human splicing factor Fox-1 bound to its consensus heptanucleotide RNA binding motif. After UV irradiation and subsequent sample preparation, the cross-linked products were analyzed by mass spectrometry (MS) to pinpoint which amino acids of the RRM cross-link to which ribonucleotides. To precisely identify the cross-linking nucleotide, the RNA motifs were synthesized with different isotopic labelling schemes, in which one of the seven nucleotides was labelled with 13C atoms. The MS signal of a given cross-link comprising a labeled nucleotide shows a defined mass shift with that of an unlabelled equivalent, which allows for the unambiguous identification of the involved nucleotide.
The surprising result for the Fox-1 RRM “protein-side” was that a cross-link can only occur if there is a Pi-stacking between an aromatic amino acid and guanosine or uridine. Moreover, this Pi-stacking requirement applies not only for cross-links to the stacked aromatic amino acid but also for its neighboring amino acids. “Discovering the Pi-stacking’s importance was an eye-opening moment and changed course of the project”, states Jonathan Hall, last author of the study published in Nature Communications.
Subsequently, the researchers looked at three published large-scale MS cross-linking datasets. Excluding cross-links to sulphur-containing amino acids, and using structural data, they looked at the percentage of cross-links in the proximity of an aromatic amino acid that is Pi-stacked to a nucleotide. In the three datasets, 42, 52 and 78 % of the cross-links involved a Pi-stacked aromatic amino acid in proximity, validating the team’s findings.
Detailed analysis of the structure-reactivity relationships of other RNA-protein complexes led to an equally surprising discovery on the “RNA-side”. The MS data for one particular histidine showed cross-linking to uracil (U), while the 3D structure indicated Pi-stacking to cytidine (C). The most likely explanation for this discrepancy was that hydrolysis of the Pi-stacking cytidine to uridine occurs during or after cross-linking. Re-inspecting data from the Fox-1 system suggested that C to U conversions can be widespread, suggesting that not only guanosine and uridine, but also cytidine can participate in UV cross-linking reactions. The unexpected finding of the C to U conversion on the “RNA-side,” as well as the importance on the “protein-side” for Pi-stacked aromatic amino acids, should be kept in mind for analyzing CLIR-MS and CLIP data.
The publication describing these findings was published in Nature Communications. “We gained a lot of insights through this project but could not determine the chemical structure of the cross-linking products. Also, extending this study to sulfur-containing amino acids would contribute further important insights into this process,” says Anna Knörlein, first author of the study.
Publication:
Knörlein et al. (2022) Nature Communications 13(1), 2719 (Open Access)
Text: Dominik Theler