Translating Time
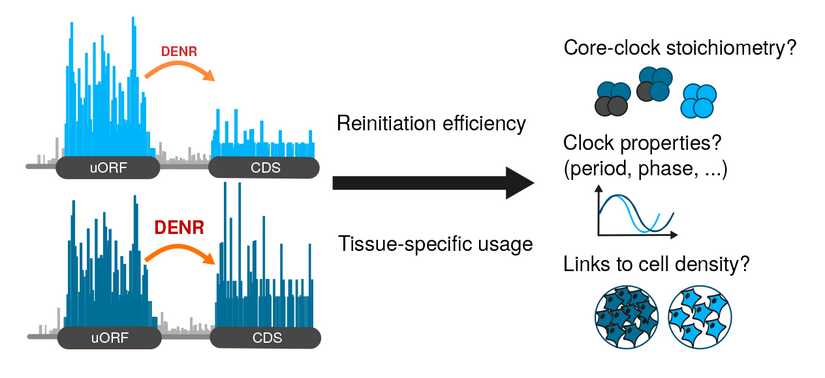
The Gatfield Lab has tackled yet another question of how posttranscriptional regulation interconnects with rhythmic gene expression driven by the circadian clock. This time, their interest was in upstream open reading frames (uORFs). They uncovered a novel mechanism how a uORF – and its capacity to trigger translation reinitiation – defines the N-terminus and the levels of the CLOCK protein. As an entry point, PhD student Violeta Castelo-Székely used fibroblast cells with a lossoffunction of DENR, which is a noncanonical initiation factor that was recently associated with reinitiation both in Drosophila and in HeLa cells. The aim of the experiment was to identify rules that explain why certain uORFs trigger DENR-dependent reinitiation while others do not. To identify DENR-regulated translation events transcriptome- wide, the researchers applied ribosome profiling to DENR-deficient NIH3T3 cells. By this method, they uncovered 240 endogenous transcripts with altered translation rate, and extracted 5’ UTR features predictive of DENR dependence.
David Gatfield explains that there are some enigmas still to be resolved when it comes to circadian rhythms and biological clocks. „Beyond the welldescribed transcriptional feedback loops that are at the basis of circadian timekeeping, we know that there are a number of unexplained features that most likely involve regulation at the RNA level.“ The researchers suspected translation to play a role, as an additional level in the regulatory network. „Already a couple of years back the hypothesis to be tested was: The translational level is important for the function of the circadian clock.“ At the time, they had investigated this hypothesis from various angles and also observed that knocking down DENR led to circadian period shortening. In the absence of DENR, the clock was ticking considerably faster, the cells lived soto- speak by a 22–23 hour day, rather than the correct 24 hours. However, which particular clock transcripts were actually regulated by DENR? The current study now uncovers that Clock itself is translationally controlled through uORFs and DENR. It turns out that Nature has devised a surprisingly complicated translational landscape in the Clock 5’ UTR: a highly translated uORF that overlaps the annotated CLOCK coding sequence in a different frame thus triggers DENR-dependent reinitiation from a downstream, alternative start codon.
“I see the study as part of a larger effort to bring some order and understanding into the current zoo of mechanisms that uORFs appear to be able to act by” explains David Gatfield. It is poorly understood why certain uORFs elicit NMD, while others trigger translation reinitiation or are subject to leaky scanning. “But of course, right now we are particularly interested in understanding this concrete, novel regulation of Clock and its importance for circadian functions and physiology in vivo”. In which physiological situation is this reinitiation event regulated? Could it serve to uncouple CLOCK protein abundance from mRNA levels in specific cell types? DENR was originally identified because its expression increased in cultured cells at high density, which led to its name Density Regulated Protein. First links between cell density and circadian rhythmicity have indeed been described in recent years, and the group considers it an interesting question for further research to assess whether CLOCK uORFs and DENR play roles in determining densitydependent differences of circadian rhythms.
As a side note, Gatfield sees the work as a good example for fruitful collaborations within the NCCR framework, in this case with the associate member Steve Pascolo at the University of Zürich. He also stresses that an important contribution to the study was made possible through an NCCR mobility grant that allowed Violeta conduct important experiments in the lab of Jernej Ule in London.
Castelo-Szekely V. et. al. (2019) Nucleic Acids Res 2019 Jun 4; 47(10):5193–5209
Text: Roland Fischer