Turbocharger for the cell machinery
That‘s how we‘ve learned the molecular-biological narrative in school, in its classic simple form: Gene-RNA-protein. DNA is transcribed into RNA and the RNA serves as a kind of punch card in the ribosomes - the proteins are assembled by the machinery according to the DNA template. In recent decades, however, biologists have realised that this scheme is far too simple, in particular with regard to the role of RNA. More and more RNA has been found bearing no code, i.e. no protein instructions. Today, it is assumed that in most living organisms the majority of RNA produced is actually "non-coding". In humans, noncoding RNA makes up an amazing 98 percent of RNA. Why is so much RNA transcribed not serving the "classical" purpose? As simple as the question is, it still offers plenty of surprising answers.
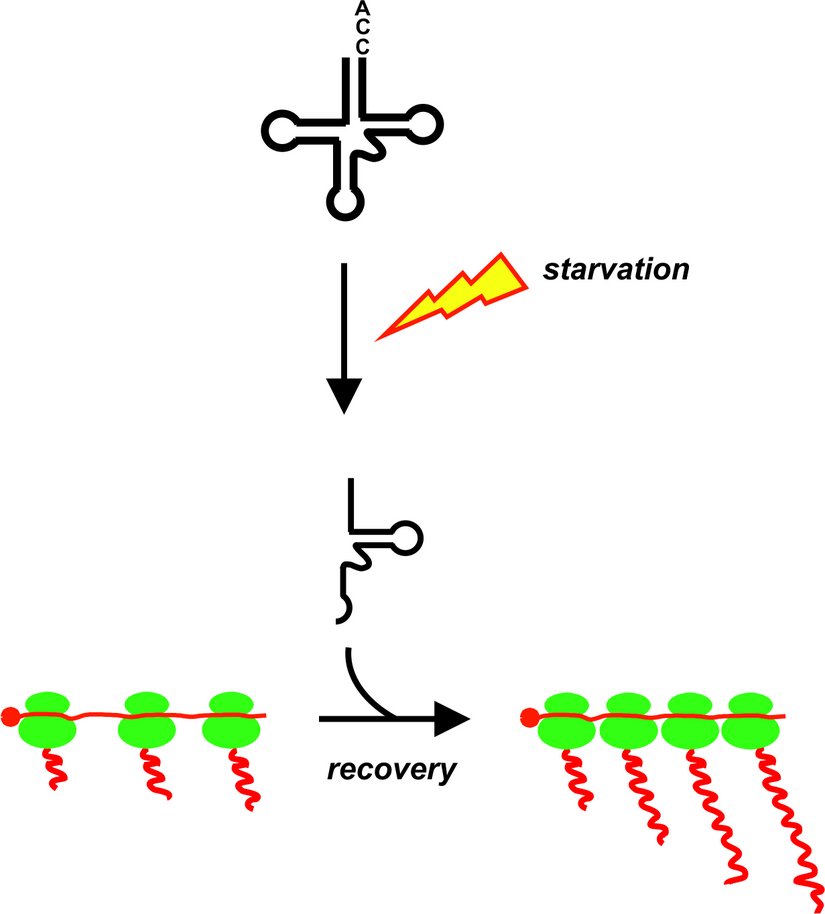
As reported in Nature Communications, Researchers from the Polacek and the Schneider groups of the University of Bern have discovered a new molecular regulatory mechanism in the unicellular parasites Trypanosoma brucei never described before. Trypanosomes, parasitic protozoa responsible for sleeping sickness, are known for their unique molecular biological apparatus. In the absence of extensive transcription control mechanisms the parasite crucially depends on translation regulation to orchestrate gene expression. However, molecular insight into regulating protein biosynthesis is sparse. The Polacek and the Schneider groups analyzed the small non-coding RNA (ncRNA) interactome of ribosomes in T. brucei during different growth conditions and life stages. Ribosome-associated ncRNAs have recently been recognized as unprecedented regulators of ribosome functions. The researchers have identified one especially intriguing ncRNA, the tRNAThr 3' half. It is produced during nutrient deprivation and becomes one of the most abundant tRNA-derived RNA fragments (tdRs). tRNAThr halves associate with ribosomes and polysomes and, once starvation conditions ceased, stimulate translation by facilitating mRNA loading during stress recovery.
These findings astonished the researchers because until now only the opposite function of non-coding RNA had been known, acting as inhibitors for the cell apparatus. During stress, ncRNA molecules attach to ribosomes, as if pushing the emergency stop button of the protein machinery. When nutrients become scarce or environmental conditions become especially challenging, the entire assembly line is shut down, saving time for the cell. ncRNA molecules are predestined for such a regulatory mechanism - they are produced within fractions of minutes and can thus trigger a fast reaction of the cell. But an acceleration of production? This irritated the researchers in two respects: Firstly, it is not immediately clear what the purpose of such a regulation might be, and secondly, it is much more difficult to come up with an intuitive mechanism for such a mode of operation. "We knew of inhibitors, which typically block important binding sites," said Norbert Polacek, the head of the research group. He believes that the ncRNA fragment has the effect of bringing the ribosomes up to full production capacity without delay as soon as, for example, sufficient nutrients are available again - Polacek calls it a " kick start " for the cell.
Blocking or depleting the endogenous tRNAThr halves mitigates this stimulatory effect both in vivo and in vitro. T. brucei and its close relatives lack the well-described mammalian enzymes for tRNA half processing, thus hinting at a unique tdR biogenesis in these parasites. The exact mechanism remains unclear, however, and Polacek believes this opens up an interesting field for further research. Furthermore, the findings widen the understanding of the regulatory potential of tdRs in general, as compared to other small ncRNA regulators. The researchers do not exclude the possibility that the T. brucei tRNAThr 3' half has additional biological roles in the parasite beyond translation control. They find it „astounding“ that the “precursor” molecule of tdRs, genuine tRNA that is, has basically one major cellular role as substrate for the protein synthesis machinery, while processing products thereof are functionally so heterogeneous. Thus post-transcriptional cleavage events can generate novel regulatory molecules thereby further increasing the complexity of cellular RNomes in general and expanding tRNA biology in particular.
Fricker R. et al. (2019) Nature Communications 10(1),118 (open access)
Text: Roland Fischer