Still conFUSed?
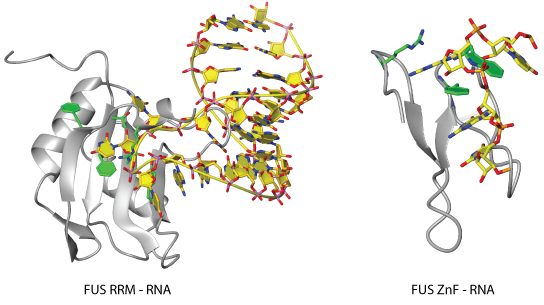
Fused in Sarcoma (FUS) is an RNA binding protein associated with several neurodegenerative diseases. RNA binding has been suggested to be crucial for FUS function and recent research has shown that FUS has the intrinsic ability to bind many RNAs without substantial differences in binding affinity. The actual mode of nucleic acid binding has been elusive, so what exactly determines the FUS interactome in vivo has become one of the big unanswered questions in the RNA field. Finally, Loughlin et al. from the Allain group (Institute of Molecular Biology and Biophysics, D-BIOL, ETH Zürich) managed to solve the solution structure of FUS bound to RNA, revealing a sequence-specific recognition for a GGU motif and an unusual shape recognition of a stem loop by two separate domains.
This is not only interesting on a theoretical level: FUS plays an important role in regulating genetic messengers and the interaction of different proteins. FUS mutations lead to FUS accumulations in the cytoplasm. The two neurodegenerative diseases amyotrophic lateral sclerosis (ALS) and fronto-temporal lobar degeneration (FTLD) show neuropathological protein aggregates containing FUS, and it is hypothesized that mis-regulation of RNA processing could play a major role in these diseases.
Transcriptomics studies have already indicated that FUS binds a large variety of RNA motifs, suggesting that FUS RNA binding can only be explained with a complex pattern. The findings of Loughlin et al. finally shed some light on the binding mode of FUS. With the help of colleagues from the Department of Chemistry and Biochemistry of the University of Bern (Mühlemann and Ruepp's labs) and the Institute of Molecular Life Sciences of the University of Zürich (Polymenidou's lab), the structure solved by the Allain group that revealed a bipartite binding mode of RNA via its RRM and zinc-finger (Znf) could be functionally validated in cell-based assays.
The ZnF provides sequence specificity to FUS, whereas the FUS RRM binds stem-loop RNAs in an unusual manner and with highly degenerate specificity. The structure of the FUS RRM bound to the stem-loop RNA reveals three individual binding pockets on the betasheet surface, as expected for an RRM. However, the path taken by these nucleotides is unusual, with the three nucleotides forming a tight turn rather than a straight line. Most contacts are of non-sequence-specific nature, with hydrophobic interactions and contacts to the phosphate backbone.
RNA binding by FUS is important in facilitating efficient liquid-liquid phase separation into membrane-less compartments like liquid droplets, the aging of which could lead to aggregation of FUS in ALS and FTLD patients. The modular nature of FUS RNA binding and the weak RNA binding affinity of the folded domains shown here are perfectly consistent with the weak multivalent interactions known to facilitate phase transition. This multivalent RNA binding, together with the disordered regions of FUS, are well suited to play a role in the formation of the different phases, and the RRM and ZnF have recently been shown to contribute to RNA-mediated phase separation of FUS. Furthermore, the role of RGG regions in destabilizing structured regions of RNA in addition to direct binding may further facilitate this process.
It is a complex story, indeed. The results not only open up interesting new paths for further research, but also help understand why deciphering the RNA binding mode of FUS has been so challenging. "This was a very difficult structure to solve due to the very dynamics nature of the interaction of the RRM with the RNA. But I am glad this could be achieved, thanks to the heroic effort of Dr. Fionna Loughlin (mother of two with 120% SNF support and now back to Australia) and a great collaborative effort from four NCCR groups" said Prof. Fred Allain.
Louglin F.E. et. al. (2019) Molecular Cell 73(3), 490-504.e6
Text: Roland Fischer