The Jinek lab together with the Allain, Palermo labs and scientists from Caribou Biosciences investigated the off-target activity of Cas9. In total, they determined 19 crystal structures of Cas9 in complex with on- and off-targets DNAs. The structures and additional experiments allowed them to see how mismatches and single nucleotide deletions can be accomodated by this widely used CRISPR enzyme. Their study has been published in the journal Cell.
Highlights
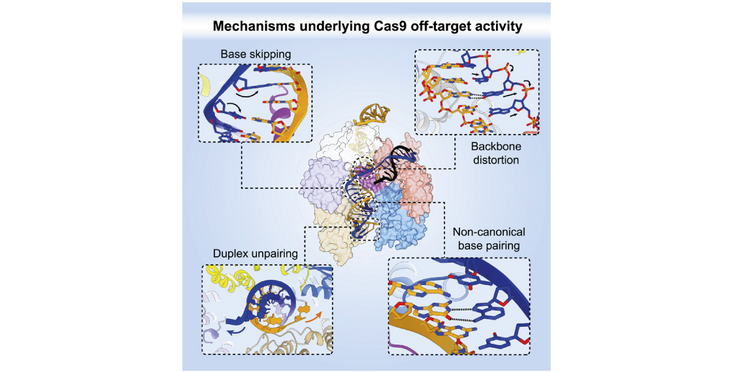
- 19 crystal structures of Cas9 bound to on- and off-target DNA substrates
- Mismatch tolerance is enabled by formation of non-canonical base pairs
- Single-nucleotide deletions are accommodated by base skipping or multiple mismatches
- PAM-distal mismatches perturb the REC2/REC3 conformational checkpoint
Abstract
The target DNA specificity of the CRISPR-associated genome editor nuclease Cas9 is determined by complementarity to a 20-nucleotide segment in its guide RNA. However, Cas9 can bind and cleave partially complementary off-target sequences, which raises safety concerns for its use in clinical applications. Here, we report crystallographic structures of Cas9 bound to bona fide off-target substrates, revealing that off-target binding is enabled by a range of noncanonical base-pairing interactions within the guide:off-target heteroduplex. Off-target substrates containing single-nucleotide deletions relative to the guide RNA are accommodated by base skipping or multiple noncanonical base pairs rather than RNA bulge formation. Finally, PAM-distal mismatches result in duplex unpairing and induce a conformational change in the Cas9 REC lobe that perturbs its conformational activation. Together, these insights provide a structural rationale for the off-target activity of Cas9 and contribute to the improved rational design of guide RNAs and off-target prediction algorithms.
Read the Publication in Cell (Open Access)
Abstract, figure and title from Pacesa et al. (2022) Cell published under a CC BY-NC-ND 4.0 license.