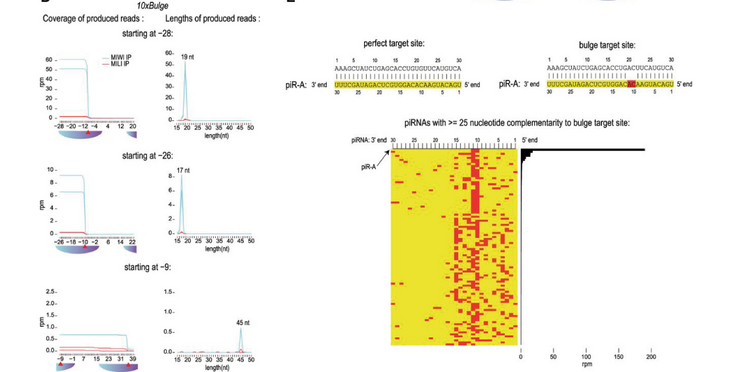
The Pillai lab found that in vivo mouse MIWI cleaves targets containing mismatches at certain positions while in vitro such targets are not sliced. In addition they discovered that not all PIWI slicing targets become mature piRNAs. Their findings were published in the article "In vivo PIWI slicing in mouse testes deviates from rules established in vitro" in RNA.
Abstract
Argonautes are small RNA-binding proteins, with some having small RNA-guided endonuclease (slicer) activity that cleaves target nucleic acids. One cardinal rule that is structurally defined is the inability of slicers to cleave target RNAs when nucleotide mismatches exist between the paired small RNA and the target at the cleavage site. Animal-specific PIWI clade Argonautes associate with PIWI-interacting RNAs (piRNAs) to silence transposable elements in the gonads, and this is essential for fertility. We previously demonstrated that purified endogenous mouse MIWI fails to cleave mismatched targets in vitro. Surprisingly, here we find using knock-in mouse models that target sites with cleavage-site mismatches at the 10th and 11th piRNA nucleotides are precisely sliced in vivo. This is identical to the slicing outcome in knock-in mice where targets are base-paired perfectly with the piRNA. Additionally, we find that pachytene piRNA-guided slicing in both these situations failed to initiate phased piRNA production from the specific target mRNA we studied. Instead, the two slicer cleavage fragments were retained in PIWI proteins as pre-piRNA and 17–19 nt by-product fragments. Our results indicate that PIWI slicing rules established in vitro are not respected in vivo, and that all targets of PIWI slicing are not substrates for piRNA biogenesis.
Read the Publication in RNA (Open Access)
Abstract, figure and title from Dowling et al (2023) RNA published under a CC BY 4.0 license.