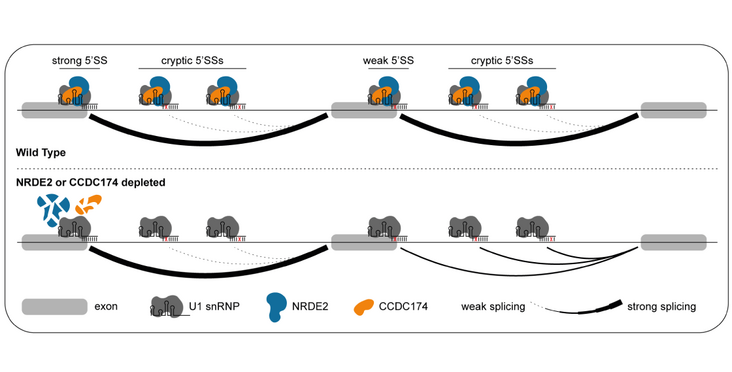
The Bühler lab by the use of their novel method BLIP-seq could shed identify two factors that promote the splicing of weak 5' splice sites which are not targeted efficiently by U1 snRNP alone. The two identified factors NRDE2 (Nuclear RNAi defective-2) and CCDC174 (Coiled-Coil Domain Containing 174), bind in mouse embryonic stem cells the 5' splice sites and directly to U1 snRNA, and promote as non-canonical splicing factors the proper splicing of hundreds of genes. Their findings have been published in the article "Mouse Nuclear RNAi-defective 2 Promotes Splicing of Weak 5' Splice Sites " in the RNA journal.
Abstract
Removal of introns during pre-mRNA splicing, which is central to gene expression, initiates by base pairing of U1 snRNA with a 5' splice site (5'SS). In mammals, many introns contain weak 5'SSs that are not efficiently recognized by the canonical U1 snRNP, suggesting alternative mechanisms exist. Here, we develop a cross-linking immunoprecipitation coupled to a high-throughput sequencing method, BCLIP-seq, to identify NRDE2 (Nuclear RNAi defective-2) and CCDC174 (Coiled-Coil Domain-Containing 174) as novel RNA-binding proteins in mouse ES cells that associate with U1 snRNA and 5'SSs. Both proteins bind directly to U1 snRNA independently of canonical U1 snRNP specific proteins, and they are required for the selection and effective processing of weak 5'SSs. Our results reveal that mammalian cells use non-canonical splicing factors bound directly to U1 snRNA to effectively select suboptimal 5'SS sequences in hundreds of genes, promoting proper splice site choice and accurate pre-mRNA splicing.
Read the Article in RNA (Open Access)
Abstract, figure and title from Flemr et al RNA (2023) published under a CC BY 4.0 license.