Review by the Johnson lab on "Designing libraries for pooled CRISPR functional screens of long noncoding RNAs" published in Mammalian Genomes.
Abstract
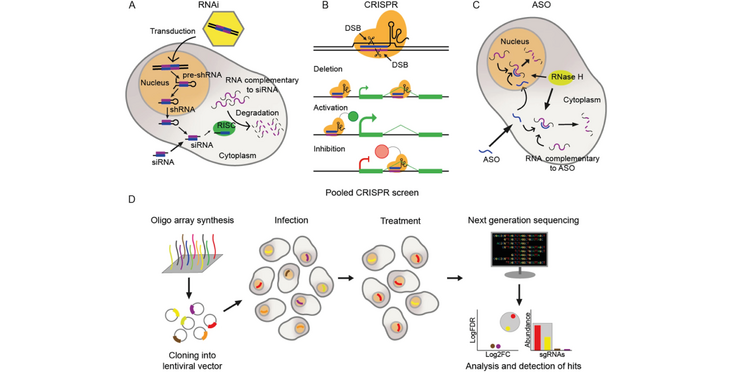
Human and other genomes encode tens of thousands of long noncoding RNAs (lncRNAs), the vast majority of which remain uncharacterised. High-throughput functional screening methods, notably those based on pooled CRISPR-Cas perturbations, promise to unlock the biological significance and biomedical potential of lncRNAs. Such screens are based on libraries of single guide RNAs (sgRNAs) whose design is critical for success. Few off-the-shelf libraries are presently available, and lncRNAs tend to have cell-type-specific expression profiles, meaning that library design remains in the hands of researchers. Here we introduce the topic of pooled CRISPR screens for lncRNAs and guide readers through the three key steps of library design: accurate annotation of transcript structures, curation of optimal candidate sets, and design of sgRNAs. This review is a starting point and reference for researchers seeking to design custom CRISPR screening libraries for lncRNAs.
Read the Article in Mammalian Genome (Open Access)
Abstract, figure and title from Pulido-Quetglas and Johnson (2021) Mammalian Genome published under a CC BY 4.0 license.