The Allain lab (ETH Zurich) together with the Sponer (Czech Academy of Sciences, Brno, Czech Republic) and Zavolan (University of Basel) labs joined forces to gain insight into the RNA binding mode of RRM1 of SRSF1. Based on the complex structure, it was possible to engineer by mutating a single amino acid in RRM1 a SRSF1 variant, which leads to inclusion of exon 7 of the SMN2 gene, which is the molecular mechanisms of action of two drugs approved for the treatment of Spinal Muscular Atrophy. Their collaborative findings have just been published in Nature Communications.
Abstract
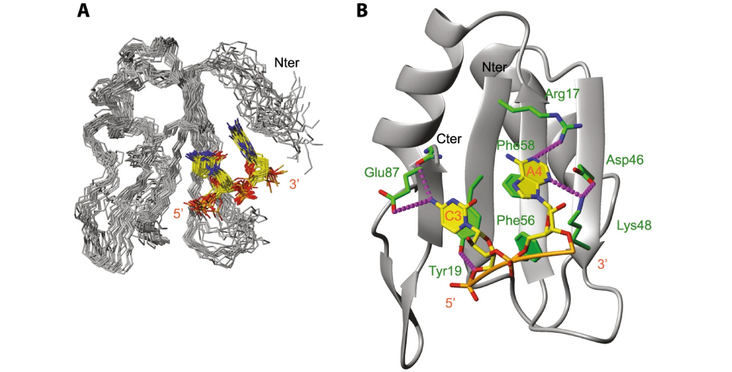
The human prototypical SR protein SRSF1 is an oncoprotein that contains two RRMs and plays a pivotal role in RNA metabolism. We determined the structure of the RRM1 bound to RNA and found that the domain binds preferentially to a CN motif (N is for any nucleotide). Based on this solution structure, we engineered a protein containing a single glutamate to asparagine mutation (E87N), which gains the ability to bind to uridines and thereby activates SMN exon7 inclusion, a strategy that is used to cure spinal muscular atrophy. Finally, we revealed that the flexible inter-RRM linker of SRSF1 allows RRM1 to bind RNA on both sides of RRM2 binding site. Besides revealing an unexpected bimodal mode of interaction of SRSF1 with RNA, which will be of interest to design new therapeutic strategies, this study brings a new perspective on the mode of action of SRSF1 in cells.
Read the Publication in Nature Communications (Open Access)
Abstract and figure from Cléry et al. (2021) Nature Communications published under CC BY 4.0 license.