The Zavolan and van Nimwegen groups developed the bioinformatical method RCRUNCH as an end-to-end solution for the analysis of CLIP data. With their new method, they analyzed the ENCODE eCLIP data to compile a compendium of RNA-binding protein binding sites and motifs. Their method and results have been published in the article "Improved analysis of (e)CLIP data with RCRUNCH yields a compendium of RNA-binding protein binding sites and motifs" in Genome Biology.
Abstract
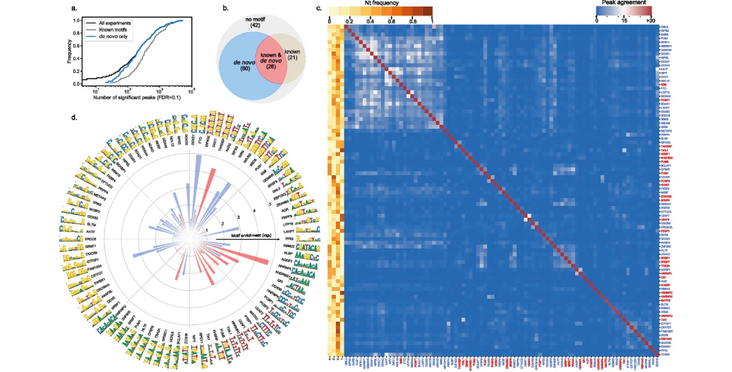
We present RCRUNCH, an end-to-end solution to CLIP data analysis for identification of binding sites and sequence specificity of RNA-binding proteins. RCRUNCH can analyze not only reads that map uniquely to the genome but also those that map to multiple genome locations or across splice boundaries and can consider various types of background in the estimation of read enrichment. By applying RCRUNCH to the eCLIP data from the ENCODE project, we have constructed a comprehensive and homogeneous resource of in-vivo-bound RBP sequence motifs. RCRUNCH automates the reproducible analysis of CLIP data, enabling studies of post-transcriptional control of gene expression.
Read the Publication in Genome Biology (Open Access)
Abstract, figure and title from Katsantoni et al (2023) Genome Biology published under a CC BY 4.0 license.