The lab of Jonathan Hall in collaboration with Mihaela Zavolan's lab investigated the targetome of two miRNAs using a new class of miR-CLIP probes. They found that the two isoforms miR-124 have a different inhibitory effect on a given target. Their findings have been published in an article entitled "MiR-CLIP reveals iso-miR selective regulation in the miR-124 targetome" in Nucleic Acids Research.
Abstract
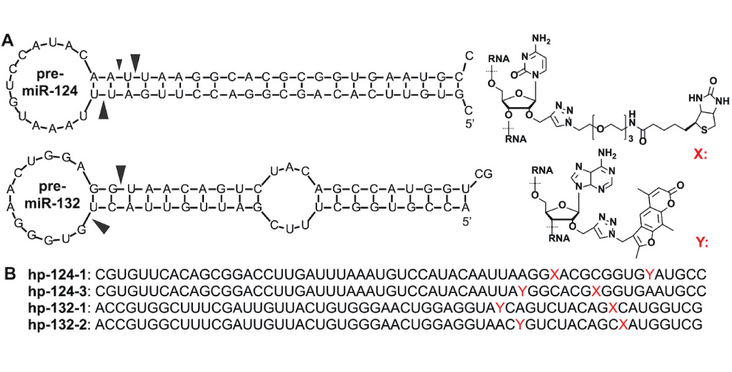
Many microRNAs regulate gene expression via atypical mechanisms, which are difficult to discern using native cross-linking methods. To ascertain the scope of non-canonical miRNA targeting, methods are needed that identify all targets of a given miRNA. We designed a new class of miR-CLIP probe, whereby psoralen is conjugated to the 3p arm of a pre-microRNA to capture targetomes of miR-124 and miR-132 in HEK293T cells. Processing of pre-miR-124 yields miR-124 and a 5'-extended isoform, iso-miR-124. Using miR-CLIP, we identified overlapping targetomes from both isoforms. From a set of 16 targets, 13 were differently inhibited at mRNA/protein levels by the isoforms. Moreover, delivery of pre-miR-124 into cells repressed these targets more strongly than individual treatments with miR-124 and iso-miR-124, suggesting that isomirs from one pre-miRNA may function synergistically. By mining the miR-CLIP targetome, we identified nine G-bulged target-sites that are regulated at the protein level by miR-124 but not isomiR-124. Using structural data, we propose a model involving AGO2 helix-7 that suggests why only miR-124 can engage these sites. In summary, access to the miR-124 targetome via miR-CLIP revealed for the first time how heterogeneous processing of miRNAs combined with non-canonical targeting mechanisms expand the regulatory range of a miRNA.
Read the Publication in Nucleic Acids Research (Open Access)
Website Hall Lab
Website Zavolan Lab
Figure and abstract from Wang et al. (2020) Nucleic Acids Research published under the CC BY 4.0 license.