Author summary
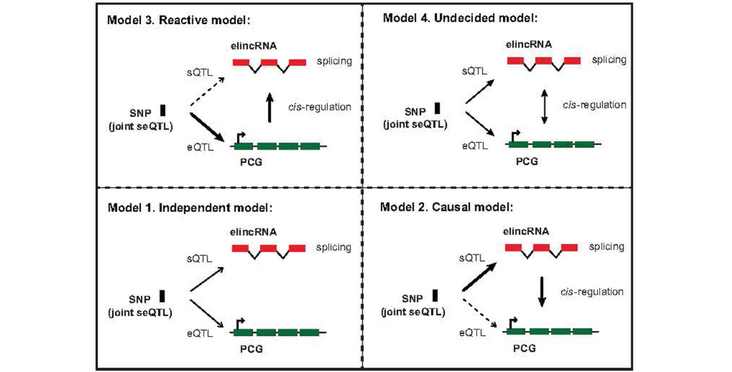
Most, if not all, active enhancers are transcribed, giving rise to a plethora of transcripts, including enhancer-associated long noncoding RNAs (elncRNAs). Changes in elncRNA levels impacts cognate enhancer activity. Recently splicing of elncRNA has also been found to associate with enhancer activity. Whether this associations reflects a contribution of elncRNA splicing to increased enhancer activity or else is simply the consequence of increased chromatin accessibility that promotes transcriptional elongation and allows for spurious splicing events remains unknown. We show that natural selection has acted, at the species and population level, to preserve DNA elements required for frequent and efficient elncRNA splicing Importantly, using a genome-wide and unbiased statistical population genomics approach, we demonstrate that elncRNA splicing is associated with cognate enhancer function, contributing to chromatin status and enhancer activity. Our results provides strong evidence that efficient elncRNA splicing contributes to enhancer activity genome-wide.
Abstract
Pervasive enhancer transcription is at the origin of more than half of all long noncoding RNAs in humans. Transcription of enhancer-associated long noncoding RNAs (elncRNA) contribute to their cognate enhancer activity and gene expression regulation in cis. Recently, splicing of elncRNAs was shown to be associated with elevated enhancer activity. However, whether splicing of elncRNA transcripts is a mere consequence of accessibility at highly active enhancers or if elncRNA splicing directly impacts enhancer function, remains unanswered. We analysed genetically driven changes in elncRNA splicing, in humans, to address this outstanding question. We showed that splicing related motifs within multiexonic elncRNAs evolved under selective constraints during human evolution, suggesting the processing of these transcripts is unlikely to have resulted from transcription across spurious splice sites. Using a genome-wide and unbiased approach, we used nucleotide variants as independent genetic factors to directly assess the causal relationship that underpin elncRNA splicing and their cognate enhancer activity. We found that the splicing of most elncRNAs is associated with changes in chromatin signatures at cognate enhancers and target mRNA expression. We provide evidence that efficient and conserved processing of enhancer-associated elncRNAs contributes to enhancer activity.
Read the Publication in PLoS Computational Biology (Open Access)
Abstract, author summary, figure and tilte from Tan and Marques (2022) PLoS Computational Biology published under a CC BY 4.0 license.