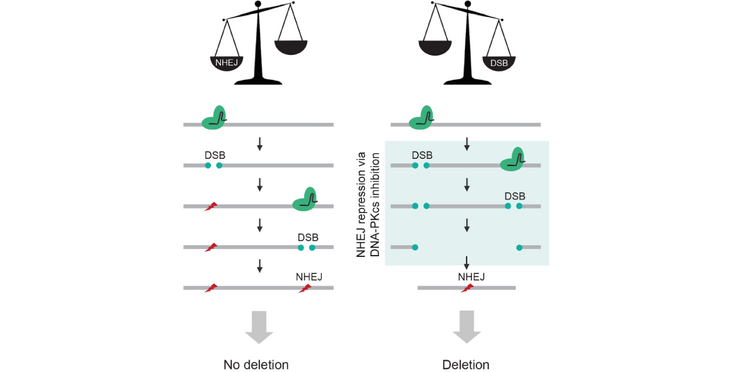
In their recent Genome Research publication the lab of Rory Johnson describes their findings how the efficiency of CRISPR deletion experiments can be boosted by a small molecule inhibitor of DNA-PKcs, which leads to slowing down of nonhomologous end-joining DNA repair.
Abstract
CRISPR-Cas9 deletion (CRISPR-del) is the leading approach for eliminating DNA from mammalian cells and underpins a variety of genome-editing applications. Target DNA, defined by a pair of double-strand breaks (DSBs), is removed during nonhomologous end-joining (NHEJ). However, the low efficiency of CRISPR-del results in laborious experiments and false-negative results. By using an endogenous reporter system, we show that repression of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs)—an early step in NHEJ—yields substantial increases in DNA deletion. This is observed across diverse cell lines, gene delivery methods, commercial inhibitors, and guide RNAs, including those that otherwise display negligible activity. We further show that DNA-PKcs inhibition can be used to boost the sensitivity of pooled functional screens and detect true-positive hits that would otherwise be overlooked. Thus, delaying the kinetics of NHEJ relative to DSB formation is a simple and effective means of enhancing CRISPR-deletion.
Read the Publication in Genome Research (Open Access)
Abstract and figure from Bosch-Guiteras et al. (2021) Genome Research published under a CC BY-NC 4.0 license