METTL16 is a m6A writer in addition to METTL3/METTL14. The lab of NCCR principal investigator Ramesh Pillai (Department of Molecular Biology, University of Geneva) published their findings on the structure, target recognition and essential function in development of this methyltransferase in a Molecular Cell article entitled “Methylation of Structured RNA by the m6A Writer METTL16 Is Essential for Mouse Embryonic Development”.
Highlights
- Structure of the METTL16 m6A writer domain with a unique N-terminal module
- N-terminal module of METTL16 is essential for charge-based binding to RNA
- METTL16 preferentially methylates adenosines within structured RNAs
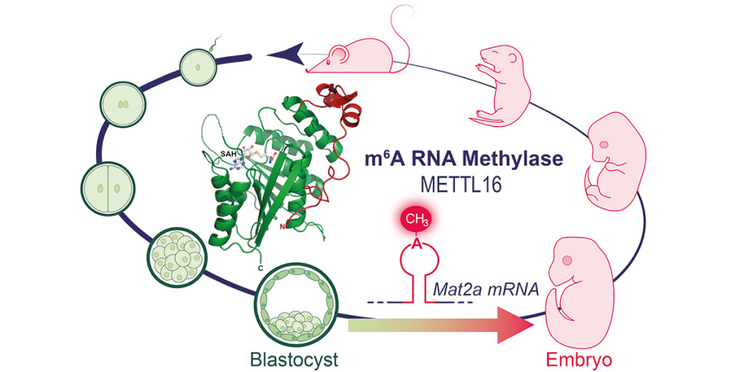
- Regulation of Mat2a mRNA by Mettl16 is essential for mouse embryonic development
Summary
Internal modification of RNAs with N6-methyladenosine (m6A) is a highly conserved means of gene expression control. While the METTL3/METTL14 heterodimer adds this mark on thousands of transcripts in a single-stranded context, the substrate requirements and physiological roles of the second m6A writer METTL16 remain unknown. Here we describe the crystal structure of human METTL16 to reveal a methyltransferase domain furnished with an extra N-terminal module, which together form a deep-cut groove that is essential for RNA binding. When presented with a random pool of RNAs, METTL16 selects for methylation-structured RNAs where the critical adenosine is present in a bulge. Mouse 16-cell embryos lacking Mettl16 display reduced mRNA levels of its methylation target, the SAM synthetase Mat2a. The consequence is massive transcriptome dysregulation in ?64-cell blastocysts that are unfit for further development. This highlights the role of an m6A RNA methyltransferase in facilitating early development via regulation of SAM availability.
Read the Publication in Molecular Cell (Open Access)
Graphical abstract, highlights and summary from Mendel et al. (2018) Molecular Cell, 71, 1-15 published under the CC BY 4.0 license.